| Name | Format | Cat No. | Price | |
|---|---|---|---|---|
| UDP-Azide-Glucose | 1 vial | 55020 | $315 | Buy |
Hydroxymethyl Collector™ Data on 5-hmC Enrichment
Hydroxymethyl Collector Range of Detection
Hydroxymethyl Collector is designed for locus-specific anlaysis of 5-hmC by real time PCR. We recommend using 5 ng - 2.5 µg fragmented genomic DNA per enrichment reaction.
Hydroxymethyl Collector–Seq is deisgned for genome-wide analysis of 5-hmC with Next-Generation sequencing. We recommend using 1 µg - 10 µg fragmented genomic DNA per enrichment reaction, with 5 µg as the suggested starting quantity. Samples containing very low levels of 5-hmC may require larger quantities of DNA. Suggested input amounts include 8-10 µg from stem cells, 20 µg from cancer tissues, 1-3 µg from brain and 5-10 µg from normal tissue.
To determine the range of detection for Hydroxymethyl Collector–Seq, various quantities of mouse and human tissues were assayed in the Hydroxymethyl Collector–Seq Kit. The enriched 5-hydroxymethylcytosine DNA fragments were analyzed by real time PCR using species-specific and tissue-specific PCR primer sets. The amount of enriched 5-hmC DNA was plotted as fold enrichment. From this analysis it was determined that Hydroxymethyl Collector–Seq is sensitive over the range of 1 - 10 µg fragmented genomic DNA.

Figure 1: Sensitivity of the Hydroxymethyl Collector–Seq Kit for enrichment of 5-hmC DNA.
Human and Mouse genomic DNA were assayed in the Hydroxymethyl Collector–Seq Kit at various quantities. Enriched DNA was analyzed by qPCR using species-specific and tissue-specific PCR primers. The enriched DNA was plotted as fold enrichment.
To determine the range of detection for Hydroxymethyl Collector, various quantities of mouse brain DNA were assayed in the Hydroxymethyl Collector Kit. The enriched 5-hydroxymethylcytosine DNA fragments were analyzed by real time PCR across multiple loci. The amount of enriched 5-hmC DNA was plotted as a percentage of the starting material. From this analysis it was determined that Hydroxymethyl Collector is sensitive over the range of 5 ng - 2.5 µg fragmented genomic DNA. For downstream analysis requiring larger inputs, e.g. microarray, it may be necessary to pool multiple enrichment reactions or perform whole-genome amplification following 5-hmC DNA enrichment.
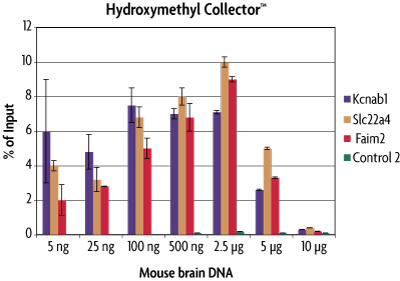
Figure 2: Sensitivity of the Hydroxymethyl Collector Kit for enrichment of 5-hmC DNA.
Mouse brain DNA was assayed in the Hydroxymethyl Collector Kit at various quantities. Enriched DNA was analyzed by qPCR across multiple loci. The enriched DNA was plotted as a percentage of starting material. This graph shows that Hydroxymethyl Collector has a preferred range of detection from 5 ng to 2.5 µg. Larger DNA quantities are not efficiently captured due to the balance of available enzyme and reagents for efficient glucosylation and biotinylation reactions. The Control 2 primer set serves as a negative control in this experiment.
Enrichment Efficiency of Hydroxymethyl Collector using the 5-hmC Control DNA
The Hydroxymethyl Collector Kit includes a 5-hmC positive control DNA, MseI digested human genomic DNA and PCR primers that can be used to determine the efficiency of the enrichment reactions. The 5-hmC Control DNA consists of a 338 base pair DNA fragment derived from the APC gene. The 5-hmC control DNA has been modified so that 25% of the cytosine residues contain 5-hydroxymethylcytosine. When the positive control fragment is spiked into human genomic DNA and enriched, the included APC PCR primers can be used to confirm the enrichment efficiency.
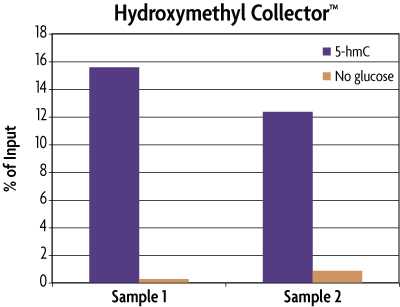
Figure 3: Enrichment of 5-hmC Control DNA included in the Hydroxymethyl Collector Kit.
MseI digested human genomic DNA (500 ng) was spiked with 25 pg of the included 5-hmC Control DNA in duplicate. These samples were then processed using the Hydroxymethyl Collector Kit to enrich for 5-hydroxymethylcytosine residues in either the presence (5-hmC) or absence (No glucose) of the UDP-Azide-Glucose donor. Eluted DNA was purified and tested using real time PCR with the included APC PCR primer mix to screen for the enrichment of the 5-hmC Control DNA. The DNA was quantified against the Input standard curve and plotted as a percentage of the starting material. This graph shows that Hydroxymethyl Collector is able to enrich for 5-hmC DNA and that the enrichment only occurs in the presence of the UDP-Azide-Glucose donor. These results are provided for demonstration only.
Applications of Hydroxymethyl Collector Enrichment
Hydroxymethyl Collector–Seq was performed on 5 µg each of mouse brain, mouse kidney and mouse liver genomic DNA and submitted for Nex-Gen sequencing to identify the localization of 5-hmC within each sample.

Figure 4: Genome-wide localization of 5-hmC in mouse tissue using Hydroxymethyl Collector–Seq.
Genomic DNA was isolated form various mouse tissues. 5 µg of DNA was used in Hydroxymethyl Collector–Seq and analyzed by Next-Generation sequencing. The sequencing peaks show the localization of 5-hmC within each sample.
Human brain DNA was used in the Hydroxymethyl Collector Kit to enrich for 5-hydroxymethylcytosine DNA fragments. Enriched DNA was amplified by whole-genome amplification and hybridized to a Human tiling array in an effort to perform genome-wide 5-hydroxymethylcytosine DNA methylation analysis.
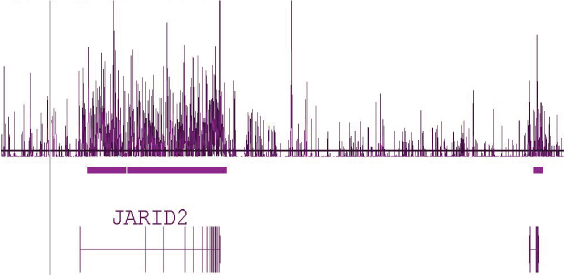
Figure 5: Human tiling array using DNA enriched with Hydroxymethyl Collector.
Human brain DNA that was enriched using the Hydroxymethyl Collector Kit was amplified by whole-genome amplification and hybridized to Affymetrix Human Tiling 2.0R Array A containing chromosomes 1 and 6. This image shows a 1.2 million base pair view of chromosome 6 where there is a clear enrichment of 5-hmC across the entire length of the JARID2 gene.
Hydroxymethyl Collector™ Method and Advantages
Advantages of the Hydroxymethyl Collector Method for 5-hmC Enrichment
- Chemical labeling ensures specific modification of 5-hmC DNA without cross-reactivity of 5-mC sites
- β-Glucosyltransferase modifies 5-hmC regardless of sequence context, enabling detection of non-CpG methylation (CpA or CpT)
- Uses dsDNA throughout the process which makes it easier to prepare libraries for downstream analysis by Next-Gen sequencing techniques
- Biotin/streptavidin binding enables more stringent binding and wash conditions to reduce non-specific background without losing the sensitivity of the assay
- Analysis is not constrained by the properties or consensus sequence of glucosyl-sensitive restriction enzymes (GSREs)
- Simple, fast procedure can be completed in less than 5 hours
- Downstream applications include individual gene analysis by PCR/qPCR or genome-wide analysis by sequencing or microarray
Flow Chart of Hydroxymethyl Collector™ Method
Below is the process of how Hydroxymethyl Collector is able to specifically modify, capture and enrich for double-stranded DNA fragments containing 5-hydroxymethylcytosine (5-hmC) residues.
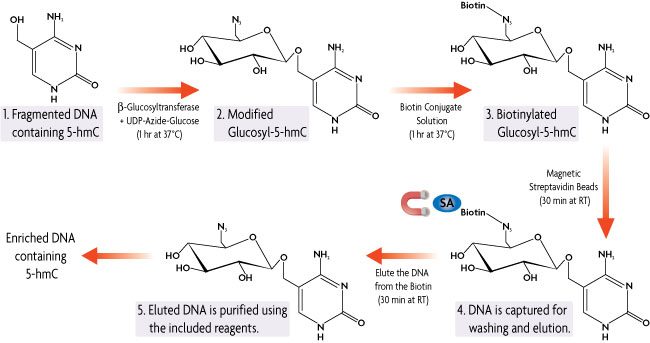
Figure 1: Hydroxymethyl Collector Method.
Hydroxymethyl Collector Kit works with fragmented (100-500 bp) double-stranded DNA. The DNA is combined with our β-Glucosyltransferase enzyme in the presence of a UDP-Azide-Glucose donor. The β-Glucosyltransferase enzyme will add the modified glucose moiety onto 5-hydroxymethylcytosine residues leaving unmethylated and 5-methylcytosine residues untouched. A chemical labeling reaction then attaches a biotin conjugate to the modified glucosyl-5-hmC DNA. Streptavidin magnetic beads are used for capture. Elution buffer is added to release the enriched DNA from the biotin linker, resulting in DNA fragments that are enriched for 5-hydroxymethylcytosine. Using the included purification reagents, the DNA is cleaned up prior to use in downstream applications.
Contents & Storage
Hydroxymethyl Collector™ contains all thre reagents needed to perform the glucosylation reaction, biotin conjugation, streptavidin capture, elution and purification of DNA containing 5-hydroxymethylcytosine. Protocols are provided for the fragmentation of sample DNA all the way through to real time PCR analysis of a locus of interest. The enriched dsDNA is compatible with Next-Generation sequencing library preparation. For added convenience, the kit also includes positive control 5-hmC DNA, which is a 338 bp sequence of the APC locus in which 25% of the cytosine residues have been hydroxymethylated. PCR primers for APC are provided to confirm the efficiency of the enrichment reactions.
Please note that the Hydroxymethyl Collector™ Kits are shipped on dry ice and contain reagents with multiple storage temperatures inside. Please store each component at the temperature indicated below. All reagents are guaranteed stable for 6 months from date of receipt when stored properly. Do not refreeze the streptavidin beads. Once thawed, store at their proper temperature. This kit includes the following components:
- β-glucosyltransferase enzyme (200 U/µl); Store at -20°C
- UDP-Azide-Glucose (3 mM); Store at -20°C
- 10X Reaction Buffer AM1; Store at -20°C
- 1 M DTT; Store at -20°C
- Biotin Conjugate Solution; Store at -20°C
- 5-hmC Control DNA (50 ng/µl); Store at -20°C
- APC PCR Primer Mix (2.5 µM); Store at -20°C
- Human genomic DNA, MseI digested (20 ng/µl); Store at -20°C (only in Hydroxymethyl Collector Catalog No. 55013)
- Streptavidin Beads; Store at 4°C
- Binding Buffer AM13; Store at 4°C
- 10X Elution Buffer AM2; Store at 4°C
- DNA Purification Binding Buffer; Store at RT
- DNA Purification Wash Buffer; Store at RT
- DNA Purification Elution Buffer; Store at RT
- ChIP Filtration Columns; Store at RT
- 3M Sodium Acetate; Store at RT
- DNA purification columns & collection tubes; Store at RT
- Bar magnet & glue dots; Store at RT

