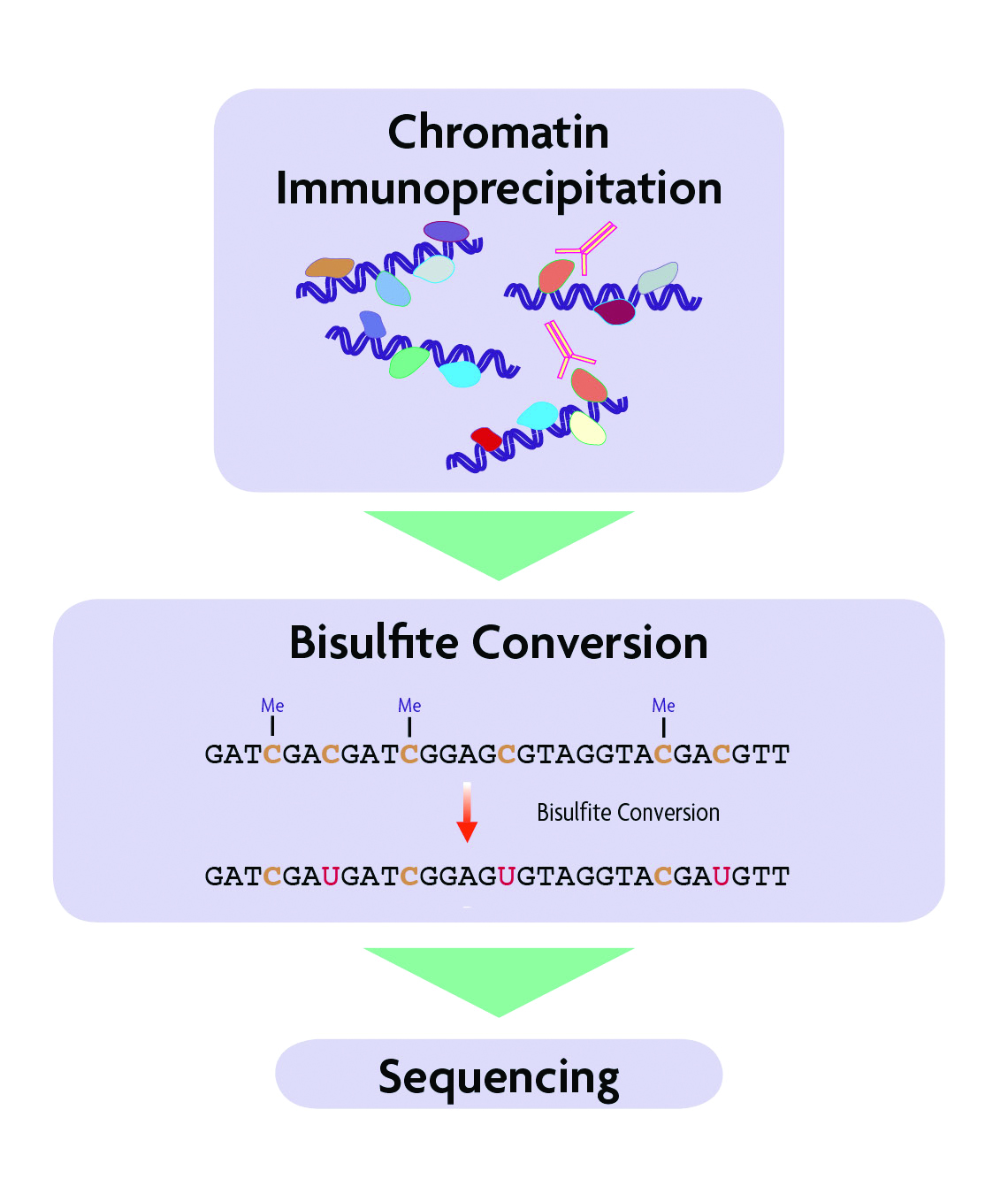
Active Motif's ChIP-Bisulfite-Sequencing (ChIP-Bis-Seq) Kit combines the target-specific selection of chromatin immunoprecipitation (ChIP) with bisulfite conversion and sequencing (Bis-Seq) to provide single nucleotide resolution of immunoprecipitated DNA. By combining the two techniques, information about both the chromatin context and the methylation profiles of each DNA strand can be evaluated, enabling better understanding of the biological context and significance regarding gene regulation, gene expression, mechanisms of chromatin modification and pathway analysis.
The ChIP-Bis-Seq Kit provides a complete and validated workflow that includes chromatin preparation and immunoprecipitation reagents, methylated adapters, sequencing library reagents and bisulfite conversion components. The ChIP-Bis-Seq Kit is compatible with Illumina® sequencing platforms.
ChIP-Bis-Seq Advantages
- Directly interrogate both chromatin and DNA methylation profiles on the same sample with higher resolution than either ChIP or bisulfite sequencing alone
- Evaluate allele-specific differences in DNA (imprinting, X-inactivation)
- Method works with histone and transcription factor antibodies
- Optimized ChIP reagents reduce background caused by non-specific binding and provide higher quality DNA for analysis
- Reduce sequencing costs by focusing on specific genomic regions of interest
- Save time and money by using our already validated ChIP-Bis-Seq workflow
THIS PRODUCT IS DISCONTINUED
| Name | Format | Cat No. | Price | |
|---|---|---|---|---|
| ChIP-Bis-Seq Kit | 10 libraries | 53048 | Discontinued | |
| High Sensitivity Chromatin Preparation | 16 rxns | 53046 | $310 | Buy |
| ChIP-Bis-Seq Kit Manual |
| High Sensitivity Chromatin Preparation Kit Manual |
| TechNote: Sonication Recommendations for ChIP |
| Epigenetics Products and Services |
| Tools for Disease Research |
How does the ChIP-Bis-Seq Kit work?

Figure 1: Flow chart of the ChIP-Bis-Seq method.
The ChIP-Bis-Seq Kit consists of three modules. The first is Active Motif’s ChIP-IT® High Sensitivity Kit which is used to immunoprecipitate chromatin associated with a protein or modification of interest in order to reduce the representation to targeted sites of the genome. The second kit module contains methylated adapters and enzymes to enable preparation of a Next-Generation Sequencing (NGS) library from the ChIP DNA. By using methylated adapters, the cytosines within the adapter sequence will remain unchanged during bisulfite conversion and retain their original sequence. The third kit module includes reagents to perform bisulfite conversion on the adapter-ligated DNA. The converted library is PCR amplified, size selected and submitted for NGS.
High Sensitivity ChIP Reagents
The ChIP-Bis-Seq Kit includes Active Motif's ChIP-IT High Sensitivity® Kit for the chromatin preparation and immunoprecipitation reactions. The ChIP-IT High Sensitivity Kit is ideal for use with challenging antibodies that do not give signal with other ChIP methods, as the method is sensitive enough to detect specific binding of even low abundance proteins. Below is a comparison of ChIP-IT High Sensitivity with other commercially available ChIP kits. ChIP was performed using untreated MCF-7 chromatin and an antibody that binds the low abundance nuclear receptor co-activator 2 (NCOA2) transcriptional co-factor. It was also performed with a negative control IgG and a negative control primer set (Active Motif Catalog No. 71001) to monitor non-specific binding. Only the ChIP-IT High Sensitivity method was able to detect NCOA2 binding at specific genomic locations (ESRRA promoter), providing approximately 20-fold higher enrichment than the negative controls. Although ESRRA shows qPCR signals using the kit from Competitor D, the enrichment levels are poor due to the high background from the negative control primer set and the non-specific binding detected with the IgG.
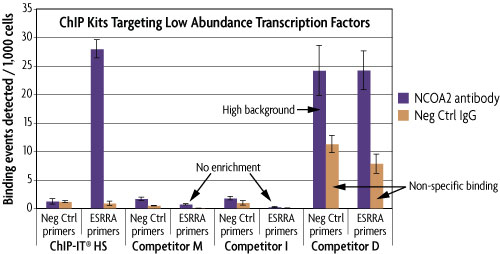
Figure 1: Comparison of ChIP kits targeting a low abundance transcription factor.
MCF-7 chromatin was prepared according to each manufacturer's recommendations for their ChIP assay from 1.5 x 106 cells. ChIP was performed with the optimal amount of chromatin as suggested by the protocol for each manufacturer using an antibody for the low abundance NCOA2 protein and a negative control IgG. Following enrichment, qPCR was performed using the ChIP-IT qPCR Analysis Kit (Catalog No. 53029) in order to normalize the data and allow for direct comparison of the results. Data is expressed as binding events detected per 1,000 cells which represents the average of the raw data triplicates adjusted for the amount of chromatin in the reaction, the resuspension volume and the primer efficiency.
Efficient Bisulfite Conversion
The ChIP-Bis-Seq Kit utilizes Active Motif's highly efficient bisulfite conversion kit. Following ChIP enrichment, the DNA undergoes end repair and the addition of A overhangs to the 3´ends of DNA fragments. Methylated adapters (compatible with Illumina seqeuncing platforms) are included in the ChIP-Bis-Seq Kit for ligation to the ChIP DNA. The methylated adapters contain methylated cytosine residues throughout the adapter sequence. This prevents conversion of the cytosines during the bisulfite conversion tratement, thereby preserving the original adapter sequence for downstream PCR amplification. During bisulfite conversion, methylated cytosines will be preserved and non-methylated cytosines will be converted into uracil with >98% efficiency. Finally, the ChIP-Bis-Seq Kit contains specialized desulfonation and purification columns that are ideal for use with bisulfite-converted, fragmented, ChIP DNA.

Figure 2: Bisulfite sequencing results of PC9 cells.
Bisulfite sequencing results for the CCND2 locus in PC9 human lung adenocarcinoma cells. Red circles are methylated CpGs and blue circles are unmethylated CpGs.
Multiple Parameter Analysis
The advantage of the ChIP-BIs-Seq Kit is that it enables direct interrogation of both chromatin and DNA methylation profiles on the same sample. It combines the enrichment properties of chromatin immunoprecipitation to look at specific protein/DNA binding interactions with the single-base resolution of bisulfite sequencing to determine the methylation profile of the DNA binding site. It may be used to evaluate allele-specific differences in DNA methylation (imprinting, X-inactivation) or to study gene regulatory effects.

Figure 3: ChIP-Bis-Seq sequencing and DNA methylation results.
This figure shows an example of sequencing results using the ChIP-Bis-Seq Kit to analyze the DNA methylation profile of DNA that was enriched using a ChIP antibody for Histone H3K27me3 in PC9 cells. The top track shows a 423 bp region of chromosome 7 from Human Hg19 genome. The next track reveals the ChIP-seq peak profile for Histone H3K27me3. The final track shows the DNA methylation profile of forward and reverse reads as analyzed by bisulfite conversion of the ChIP-enriched DNA. The blue bars represent unmethylated CpGs, while the red bars represent CpGs containing methylation.
Contents & Storage
The ChIP-Bis-Seq Kit contains separate modules for each stage of the process. One module is for chromatin preparation and immunoprecipitation, another module contains Next-Generation sequencing library reagents and a third module includes the reagents needed for bisulfite conversion. Reagents within each module may contain multiple storage temperatures. All components can be stored at -20°C prior to first use, then we recommend storing each component at the temperatures indicated below. Do not re-freeze the Protein G Agarose Beads once they have been thawed for use.
Reagents for Chromatin Preparation and Immunoprecipitation (16 reactions)
- RNase A (10 µg/µl); Store at -20°C
- Proteinase K (10 µg/µl); Store at -20°C
- Blocker; Store at -20°C
- 100 mM PMSF; Store at -20°C
- Protease Inhibitor Cocktail (PIC); Store at -20°C
- Precipitation Buffer; Store at -20°C
- Carrier; Store at -20°C
- 10X PBS; Store at -20°C
- Fixation Buffer; Store at 4°C
- Protein G Agarose Beads; Store at 4°C
- TE, pH 8.0; Store at RT
- Detergent; Store at RT
- Stop Solution; Store at RT
- Chromatin prep Buffer; Store at RT
- ChIP Filtration Columns; Store at RT
- ChIP Buffer; Store at RT
- 5 M NaCl; Store at RT
- Wash Buffer AM1; Store at RT
- Elution Buffer AM4; Store at RT
- DNA Purification Binding Buffer; Store at RT
- 3 M Sodium Acetate; Store at RT
- DNA Purification Wash Buffer; Store at RT
- DNA Purification Elution Buffer; Store at RT
- DNA Purification Columns; Store at RT
Reagents for Sequencing Library Preparation (10 libraries)
- 10X Reaction Buffer AM3; Store at -20°C
- 2X Quick Ligation Buffer; Store at -20°C
- 10X T4 DNA Ligase Buffer; Store at -20°C
- DNA Polymerase I Klenow (5 U/µl); Store at -20°C
- dNTP Mix, 10 mM each; Store at -20°C
- Klenow Fragment (3´ -5´ exo-) (5 U/µl); Store at -20°C
- T4 DNA Ligase (2,000 U/µl); Store at -20°C
- T4 DNA Polymerase (3 U/µl); Store at -20°C
- T4 Polynucleotide Kinase; Store at -20°C
- 1 mM dATP; Store at -20°C
- Methylated adapters (5 µM); Store at -20°C
- Glycogen; Store at -20°C
Reagents for Bisulfite Conversion (10 conversions)
- Conversion Reagent; Store at RT
- Hydroquinone; Store at RT
- Buffer A; Store at RT
- Buffer B; Store at RT
- DNA Binding Buffer; Store at RT
- DNA Wash Buffer; Store at RT
- DNA Elution Buffer; Store at RT
- ChIP-Bis-Seq purification columns & collection tubes; Store at RT