ChIP-Seq Spike-In Normalization Overview
How Quantitative is Traditional ChIP-Seq?
ChIP-Seq is a powerful tool for genome-wide identification of transcription factor binding sites and histone post-translational modifications, but its inherent semi-quantitative nature can potentially limit researchers' ability to compare differences between samples in some cases (described in greater detail in this webinar). However, by "spiking in" a small amount of exogenous chromatin into your samples prior to the ChIP reaction, you can normalize the signal from your experimental samples to this control in your final sequencing data. This approach allows researchers to be more confident than ever in the accuracy of their inter-sample comparisons.
How Can Spike-In Normalization Improve ChIP-Seq Analysis?
One particularly compelling example of the importance of ChIP-Seq normalization is illustrated in experiments investigating the genome-wide localization of the repressive histone mark H3K27me3 in the presence of epigenetic inhibitors. H3K27me3 has one identified methyltransferase (EZH2) responsible for catalyzing methyl group addition. However, EZH2 inhibition did not appear to significantly affect H3K27me3 levels in a ChIP-Seq experiment (top two tracks in Figure 1).
Following normalization to spike-in, the significant loss of H3K27me3 becomes apparent (bottom two tracks in Figure 1). Therefore, spike-in can reveal changes in histone modifications that would otherwise be masked by experimental artifacts.
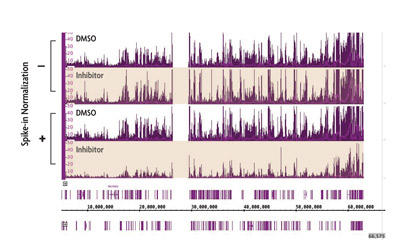
Figure 1: ChIP Normalization reveals reduction in H3K27me3 caused by an EZH2 inhibitor.
For more details, read our PLoS One publication about this spike-in normalization technique:
Egan B, Yuan C-C, Craske ML, Labhart P, Guler GD, Arnott D, et al. (2016) An Alternative Approach to ChIP-Seq Normalization Enables Detection of Genome-Wide Changes in Histone H3 Lysine 27 Trimethylation upon EZH2 Inhibition. PLoS ONE 11(11): e0166438. https://doi.org/10.1371/journal.pone.0166438
ChIP Spike-in Normalization Advantages
- Can be applied across different antibodies and samples without bias
- Spike-in Chromatin and Spike-in Antibody can be used with any ChIP kit or protocol
- Strategy works with both qPCR and ChIP-Seq analysis
How Does ChIP-Seq Spike-In Work?
ChIP normalization can be easily implemented simply by integrating our Spike-in reagents into your standard ChIP protocol using experimental chromatin and an antibody of interest. Spike-in Chromatin and Spike-in Antibody are also added, or "spiked in", to the ChIP reaction as a minor fraction of the IP reaction. Any variation introduced during the ChIP reaction will also occur with the Spike-in Chromatin. As the amount of spiked-in chromatin is consistent across all samples, a normalization factor can be created based on the spike-in signal and applied to the experimental signal.
The diagram below depicts how easily Spike-in chromatin and a Spike-in Antibody can be integrated into an existing ChIP-Seq workflow. Spike-in Chromatin and the Spike-in Antibody are added to experimental chromatin and the experimental antibody just prior to immunoprecipitation. The Spike-in Antibody recognizes a histone variant that is specific to the species of the Spike-in Chromatin (Drosophila), and the experimental antibody specifically recognizes the experimental chromatin. This enables specific detection of the Spike-in Chromatin without any significant increase in background signal. Following sequencing, reads will be mapped to their specific species. Variation introduced during the ChIP procedure will affect the Spike-in Chromatin in the same manner as the experimental chromatin, so a normalization factor can be created from the Spike-in Chromatin and applied to the experimental chromatin to normalize out technical variation and sample bias, or to monitor biological effects.
Figure 2: ChIP-Seq Normalization Workflow.
A standard ChIP reaction is set up using experimental chromatin (e.g. human) and an antibody of interest. In addition, Drosophila melanogaster chromatin is added, or "spiked-in" to each reaction as a minor fraction of the total chromatin. An antibody that recognizes the Drosophila-specific histone variant, H2Av, is added to the reaction. The Spike-in Antibody provides a mechanism to reliably pull down a small fraction of the Drosophila chromatin that is consistent across all samples. Following ChIP sequencing, the data is mapped to both the Drosophila genome and the experimental genome. A normalization factor is created for each sample based on the Drosophila tag counts. The experimental tag counts are normalized by the same factor.
ChIP Spike-In Reaction Guidelines
Each lot of Spike-in Chromatin is quantified and tested with the Spike-in Antibody. This allows us to recommend an appropriate amount of Spike-in Chromatin to use in your reaction. A minimum amount of Spike-in Chromatin is necessary to accurately normalize the sample, but Spike-in reads should take up no more than 5% of your sequencing run. The table below is an example of the recommendations made. A lot-specific data sheet accompanies each shipment, specifying the amount of Spike-in Chromatin recommended.
| Sample Chromatin | Spike-in Chromatin | Antibody of Interest | Spike-in Antibody | |
|---|---|---|---|---|
| Robust antibodies against abundant histone modifications | 25 µg | Refer to lot-specific data sheet | 4 µg | 2 µg |
| Antibodies against transcription factors, histone modifiers or low abundance histone modifications | 25 µg | Refer to lot-specific data sheet | 4 µg | 2 µg |
ChIP-Seq Spike-In Normalization Contents
Please note that the ChIP normalization reagents are available separately. Both the Spike-in Chromatin and Spike-in Antibody are required to apply the normalization strategy. Drosophila-specific qPCR primer sets are available for ChIP qPCR analysis.
Spike-in Antibody
- 50 µg Spike-in Antibody supplied at a concentration of 1 µg/µl in PBS containing 0.035% sodium azide and 30% glycerol. Spike-in Antibody is shipped at room temperature. This will not affect the stability or performance of the reagent. Upon receipt, store at -20°C. Avoid repeated freeze/thaw cycles.
Spike-in Chromatin
- Spike-in Chromatin prepared from Schneider's Drosophila Line 2 (S2) cells is provided for 15 rxns of robust histone modification antibody targets. Spike-in Chromatin is provided at a concentration of 10 ng/µl.
qPCR Primer Sets
- 400 µl Drosophila Positive Control Primer Set Pbgs is supplied at a concentration of 2.5 µM
- 400 µl Drosophila Negative Control Primer Set 1 is supplied at a concentration of 2.5 µM
ChIP-Seq Spike-In Normalization Data
Reduce the Effects of Technical Variation
The ChIP normalization strategy is ideal to correct for differences that results from sample loss, amplification bias, uneven sequencing read depth, or hand-to-hand differences between users. By utilizing the differences observed between samples with the Spike-in Chromatin, a normalization factor is created and applied to the experimental samples to normalize out the effects of technical variation.
Figure 3: ChIP Normalization of technical variation by ChIP qPCR.
A ChIP qPCR reaction was set up with 750 ng of Spike-in Chromatin added to 30 µg of human chromatin. Both Spike-in Antibody (2 µg) and a Histone H3K27me3 antibody (4 µg) were also added to the ChIP reaction. ChIP was performed according to Active Motif's ChIP-IT High Sensitivity® protocol (Catalog No. 53040), however, one sample was left as normal, one sample was given only 50% of the Protein G beads and the third sample had 50% of the IP volume removed. Enriched DNA was analyzed by qPCR using both Drosophila-specific primers and human-specific primers for each sample. The results show the effects of these sample differences on the enrichment. The Drosophila data was used to create a normalization factor for each sample. This normalization factor was then applied to each human data point. The normalized data reveals comparable ChIP qPCR results following the removal of technical variation effects, validating the spike-in strategy for normalization.
Identify Biological Differences Not Observed by Standard ChIP Analysis
By adding Spike-in Chromatin and Spike-in Antibody to standard ChIP reactions, experimental data can be normalized for sample variation. This normalization makes it easier to monitor the effects of experimental conditions, such as inhibitory compounds or mutants to reveal biological differences.

Figure 4: Normalization of biological differences in ChIP-Seq.
ChIP-Seq was performed on untreated cells and cells treated with a small molecule inhibitor of EZH2 methyltransferase. Using standard ChIP-Seq analysis (–) the differences in signal are not detected. Incorporation of the spike-in normalization strategy (+) reveals the expected decrease in H3K27me3 ChIP-Seq signal confirming the value of the normalization strategy for detecting biological changes.
Specificity of Detection
The Spike-in Chromatin consists of Drosophila melanogaster chromatin prepared from Schneider's Drosophila Line 2 (S2) cells. The Spike-in antibody recognizes a Drosophila-specific histone variant, H2Av. Because of the specificity of the Spike-in Antibody for the Spike-in Chromatin modification, there is no cross-reactivity with mammalian samples leading to reduced background signal.
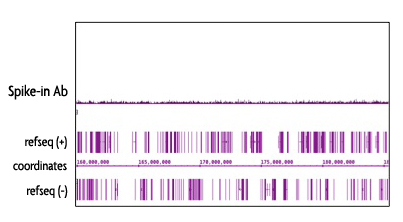
Figure 5: Specificy of the Spike-in Antibody.
The Spike-in Antibody shows minimal cross reactivity with mammalian samples. When the Spike-in Antibody was tested in ChIP-Seq with human chromatin, there is little to no signal detected. This demonstrates the specificity of the spike-in normalization strategy.
ChIP-Seq Spike-In Normalization Publications
Spike-in Antibody (Cat. No. 61686)
Search our database of customer publications that have used our Spike-in Antibody.
Spike-in Chromatin (Cat. No. 53083)
Search our database of customer publications that have used our Spike-in Chromatin.
ChIP-Seq Spike-In Normalization Documents
You might also be interested in:
| Name | Format | Cat No. | Price | |
|---|---|---|---|---|
| Spike-in Antibody | 50 µg | 61686 | $245 | Buy |
| Spike-in Chromatin | 15 rxns | 53083 | $225 | Buy |
| Drosophila Positive Control Primer Set Pbgs | 96 rxns | 71037 | $125 | Buy |
| Drosophila Negative Control Primer Set 1 | 96 rxns | 71028 | $125 | Buy |
| Drosophila Negative Control Primer Set 3 | 96 rxns | 71038 | $125 | Buy |
How does the ChIP Spike-in Normalization Strategy work?
The diagram below depicts how easily Spike-in chromatin and a Spike-in Antibody can be integrated into an existing ChIP-Seq workflow. Spike-in Chromatin and the Spike-in Antibody are added to experimental chromatin and the experimental antibody just prior to immunoprecipitation. The Spike-in Antibody recognizes a histone variant that is specific to the species of the Spike-in chromatin (Drosophila), and the experimental antibody specifically recognizes the experimental chromatin. This enables specific detection of the Spike-in Chromatin without any significant increase in background signal. Following sequencing, reads will be mapped to their specific species. Variation introduced during the ChIP procedure will affect the Spike-in Chromatin in the same manner as the experimental chromatin, so a normalization factor can be created from the Spike-in Chromatin and applied to the experimental chromatin to normalize out technical variation and sample bias, or to monitor biological effects.
Figure 1: ChIP-Seq Normalization Workflow.
A standard ChIP reaction is set up using experimental chromatin (e.g. human) and an antibody of interest. In addition, Drosophila melanogaster chromatin is added, or "spiked-in" to each reaction as a minor fraction of the total chromatin. An antibody that recognizes the Drosophila-specific histone variant, H2Av, is added to the reaction. The Spike-in antibody provides a mechnaism to reliable pull down a small fraction of the Drosophila chromatin that is consistent across all samples. Following ChIP sequencing, the the data is mapped to both the Drosophila genome and the experimental genome. A normalization factor is created for each sample based on the Drosophila tag counts. The experimental tag counts are normalized by the same factor.
ChIP Spike-in Reaction Guidelines
Each lot of Spike-in Chromatin is quantified and tested with the Spike-in Antibody. This allows us to recommend an appropriate amount of Spike-in Chromatin to use in your reaction. A minimum amount of Spike-in Chromatin is necessary to accurately normalize the sample, but Spike-in reads should take up no more than 5% of your sequencing run. The table below is an example of the recommendations made. A lot-specific data sheet accompanies each shipment, specifying the amount of Spike-in Chromatin recommended.
| Sample Chromatin | Spike-in Chromatin | Antibody of Interest | Spike-in Antibody | |
|---|---|---|---|---|
| Robust antibodies against abundant histone modifications | 25 µg | Refer to lot-specific data sheet | 4 µg | 2 µg |
| Antibodies against transcription factors, histone modifiers or low abundance histone modifications | 25 µg | Refer to lot-specific data sheet | 4 µg | 2 µg |
Reduce the Effects of Technical Variation
The ChIP Normalization Strategy is ideal to correct for differences that results from sample loss, amplification bias, uneven sequencing read depth or hand-to-hand differences between users. By utilizing the differences observed between samples with the Spike-in chromatin, a normalization factor is created and applied to the experimental samples to normalize out the effects of technical variation.
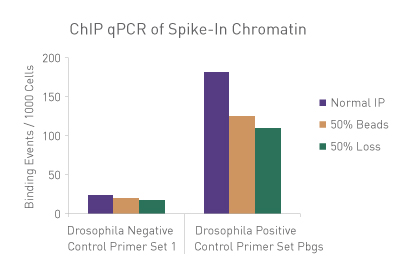
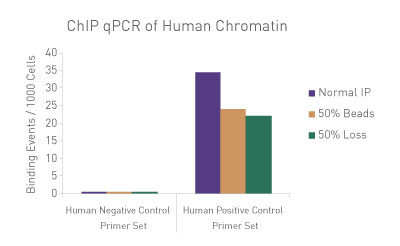
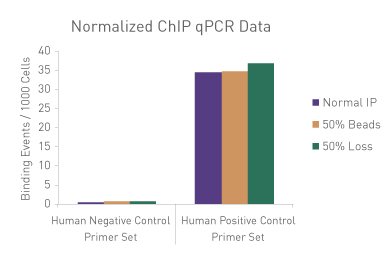
Figure 1: ChIP Normalization of technical variation by ChIP qPCR.
A ChIP qPCR reaction was set up with 750 ng of Spike-in chromatin added to 30 µg of human chromatin. Both Spike-in Antibody (2 µg) and a Histone H3K27me3 antibody (4 µg) were also added to the ChIP reaction. ChIP was performed according to Active Motif's ChIP-IT High Sensitivity protocol (Catalog No. 53040), however, one sample was left as normal, one sample was given only 50% of the Protein G beads and the third sample had 50% of the IP volume removed. Enriched DNA was analyzed by qPCR using both Drosophila-specific primers and Human-specific primers for each sample. The results show the effects of these sample differences on the enrichment. The Drosophila data was used to create a normalization factor for each sample. This normalization factor was then applied to each Human data point. The normalized data reveals comparable ChIP qPCR results following the removal of technical variation effects, validating the Spike-in strategy for normalization.
Identify Biological Differences Not Observed by Standard ChIP Analysis
By adding Spike-in Chromatin and Spike-in Antibody to standard ChIP reactions, experimental data can be normalized for sample variation. This normalization makes it easier to monitor the effects of experimental conditions, such as inhibitory compounds or mutants to reveal biological differences.

Figure 2: Normalization of biological differences in ChIP-Seq.
ChIP-Seq was performed on untreated cells and cells treated with a small molecule inhibitor of EZH2 methyltransferase. Using standard ChIP-Seq analysis (–) the differences in signal are not detected. Incorporation of the Spike-in Normalization Strategy (+) reveals the expected decrease in H3K27me3 ChIP-Seq signal confirming the value of the normalization strategy for detecting biological changes.
Specificity of Detection
The Spike-in chromatin consists of Drosophila melanogaster chromatin prepared from Schneider's Drosophila Line 2 (S2) cells. The Spike-in antibody recognizes a Drosophila-specific Histone variant, H2Av. Because of the specificity of the Spike-in antibody for the Spike-in chromatin modification, there is no cross-reactivity with mammalian samples leading to reduced background signal.

Figure 3: Specificy of the Spike-in Antibody.
The Spike-in antibody shows minimal cross reactivity with mammalian samples. When the Spike-in antibody was tested in ChIP-Seq with human chromatin, there is little to no signal detected. This demonstrates the specificity of the spike-in normalization strategy.
Contents & Storage
Please note that the ChIP Normalization reagents are available separately. Both the Spike-in Chromatin and Spike-in Antibody are required to apply the normalization strategy. Drosophila-specific qPCR primer sets are available for ChIP qPCR analysis.
Spike-in Antibody
- 50 µg Spike-in Antibody supplied at a concentration of 1 µg/µl in PBS containing 0.035% sodium azide and 30% glycerol. Spike-in Antibody is shipped at room temperature. This will not affect the stability or performance of the reagent. Upon receipt, store at -20°C. Avoid repeated freeze/thaw cycles.
Spike-in Chromatin
- Spike-in Chromatin prepared from Schneider's Drosophila Line 2 (S2) cells is provided for 15 rxns of robust histone modification antibody targets. Spike-in chromatin is provided at a concentration of 10 ng/µl.
qPCR Primer Sets
- 400 µl Drosophila Positive Control Primer Set Pbgs is supplied at a concentration of 2.5 µM
- 400 µl Drosophila Negative Control Primer Set 1 is supplied at a concentration of 2.5 µM




