RIME Services Overview
RIME (Rapid Immunoprecipitation Mass spectrometry of Endogenous proteins) is a powerful technique ideally suited for the identification of transcriptional co-factors and chromatin associated proteins.
The RIME methodology was originally described in this Cell Reports publication.
The RIME Service includes:
- Nuclear isolation and sonication.
- Immunoprecipitation.
- Purification of proteins and tryptic digestion.
- Mass spectrometry.
- Data analysis.
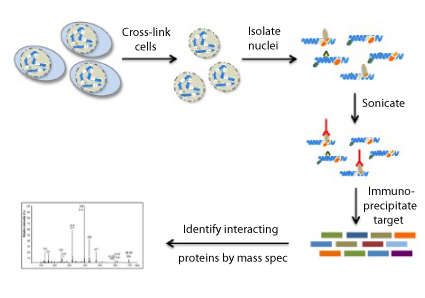
What does RIME do?
With RIME, mass spectrometry is used, along with immunoprecipitation of a target protein, to detect endogenous-interacting proteins in formaldehyde cross-linked cells.
Cross-linking advantages:
- Preserves bona fide interactions allowing for more stringent washes and minimal detection of non-specific interactions.
- Captures low-affinity interactions that are typically lost during washing.
- Captures adjacent DNA binding proteins that participate in gene regulation but do not directly interact with the targeted protein.
Benefits of RIME include:
- Identify transcriptional co-factors/co-regulators.
- Identify proteins complexed with epigenetic modifiers.
- Detect low-affinity interactions.
- Map protein interaction networks.
- Validate identified proteins by ChIP-Seq and map global co-occupancy with the RIME target protein.
- Understand how co-factors participate in differential gene regulation.
- Identify the prominent co-occurrence of transcription factor binding at adjacent sites.
What our customers are saying about us:
RIME Services Data
Figure 1: Venn Diagram of detected proteins in RIME treated MCF7 cells
RIME was performed to identify proteins that interact with the RNA polymerase machinery. Immunoprecipitations were performed with an antibody against POLR2A and two different amounts of cross-linked chromatin from MCF7 cells. Immunoprecipitated proteins were measured by mass spectrometry. The Venn diagram shows strong overlap of the detected proteins even in experiments using a 10 fold difference in starting material.
Figure 2. Detection of target protein POLR2A and potential proteins of interest
The upper panel shows the percent coverage in duplicate RIME reactions performed for POLR2A using 500ug of sample chromatin. Each experiment is assigned a grade to describe the performance of the antibody a specific sample, with >40% indicating a successful assay with very good coverage (<10% suggests that the antibody did not work well in RIME). In the bottom panel, the top 13 proteins of interest that were found to interact with POLR2A in this experiment are listed, after removing proteins present in the IgG negative control.
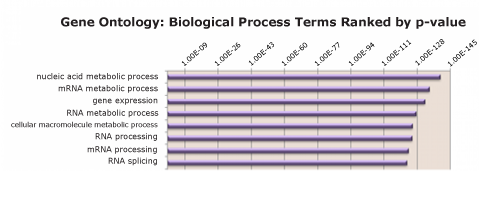
Figure 3: RIME identifies most prevalent molecular pathways among enriched proteins
PANTHER Functional Classification was performed for the list of proteins identified as interacting with POLR2A. The top eight gene ontology terms are identified and ranked by p-value.
Advanced Application: RIME and ChIP-Seq
RIME was performed using BRD4 as the target protein and from the list of interacting proteins. DNA Topoisomerase 2A was identified as an interactor, which is supported by literature demonstrating that ligand-dependent Topoisomerases have been shown to be recruited to enhancers†. Therefore, ChIP-Seq targeting BRD4 and TOP2A was performed to show co-localization. Expanding on RIME data with ChIP-Seq can help elucidate gene subsets that require different transcription factors or confirm co-localization of RIME-identified co-factors to the same genes.
Figures 4 & 5: ChIP-Seq shows co-localization of TOP2A and BRD4 binding sites.
ChIP-Seq was performed to map binding sites for the RIME-identified BRD4 interacting protein, TOP2A. TOP2A and BRD4 binding sites are shown and confirm co-localization at many sites across the genome. The top panel shows sites on chromosome 1 and the bottom panel shows sites on chromosome 3.
References
- †Puc, J. et al. Ligand-Dependent Enhancer Activation Regulated by Topoisomerase-I Activity, Cell. 2015 Oct 8;163(2):520-2.
RIME Services Publications
Search our database of customer publications that have used our RIME services.
RIME Services Documents
RIME Services Sample Submission Portal
Our online sample submission portal allows you to easily upload your service project samples and track your project status. Follow the sample submission instructions in the portal to ensure that all your samples arrive at Active Motif in the best possible condition and properly associated with your project.
You might also be interested in:
- Products (1)
- Documents (3)
- What does it do?
- RIME Data
- Advanced Application
- RIME Services Publications
| Name | Cat No. | Price | |
|---|---|---|---|
| RIME | 25067 | Get Quote | |
| Active Motif Epigenetic Services Brochure |
| RIME Services Sample Preparation |
| Epigenetic Services Citations |
With RIME, mass spectrometry is used, along with immunoprecipitation of a target protein, to detect endogenous-interacting proteins in formaldehyde cross-linked cells.
Advantages of cell cross-linking include:
- Preserves bona fide interactions allowing for more stringent washes and minimizing the detection of non-specific interactions.
- Captures low-affinity interactions that are typically lost during washing.
- Captures adjacent DNA binding proteins that participate in gene regulation but do not function through direct interaction with the targeted protein.
Benefits of RIME include:
- Identify transcriptional co-factors/co-regulators.
- Identify proteins complexed with epigenetic modifiers.
- Detect low-affinity interactions.
- Map protein interaction networks.
- Validate identified proteins by ChIP-Seq and map global co-occupancy with the RIME target protein.
- Understand how co-factors participate in differential gene regulation.
- Identify the prominent co-occurrence of transcription factor binding at adjacent sites.
Figure 1: Venn Diagram of detected proteins in RIME treated MCF7 cells
RIME was performed to identify proteins that interact with the RNA polymerase machinery. Immunoprecipitations were performed with an antibody against POLR2A and two different amounts of cross-linked chromatin from MCF7 cells. Immunoprecipitated proteins were measured by mass spectrometry. The Venn diagram shows strong overlap of the detected proteins even in experiments using a 10 fold difference in starting material.
Figure 2. Detection of target protein POLR2A and potential proteins of interest
The upper panel shows the percent coverage in duplicate RIME reactions performed for POLR2A using 500ug of sample chromatin. Each experiment is assigned a grade to describe the performance of the antibody a specific sample, with >40% indicating a successful assay with very good coverage (<10% suggests that the antibody did not work well in RIME). In the bottom panel, the top 13 proteins of interest that were found to interact with POLR2A in this experiment are listed, after removing proteins present in the IgG negative control.
Advanced Application: RIME and ChIP-Seq
RIME was performed using BRD4 as the target protein and from the list of interacting proteins. DNA Topoisomerase 2A was identified as an interactor, which is supported by literature demonstrating that ligand-dependent Topoisomerases have been shown to be recruited to enhancers†. Therefore, ChIP-Seq targeting BRD4 and TOP2A was performed to show co-localization. Expanding on RIME data with ChIP-Seq can help elucidate gene subsets that require different transcription factors or confirm co-localization of RIME-identified co-factors to the same genes.
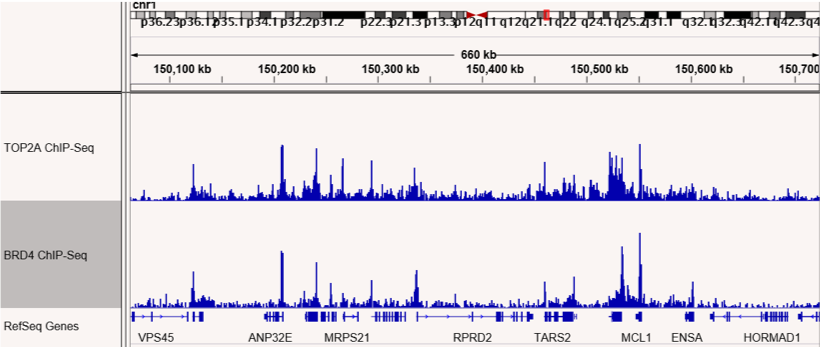
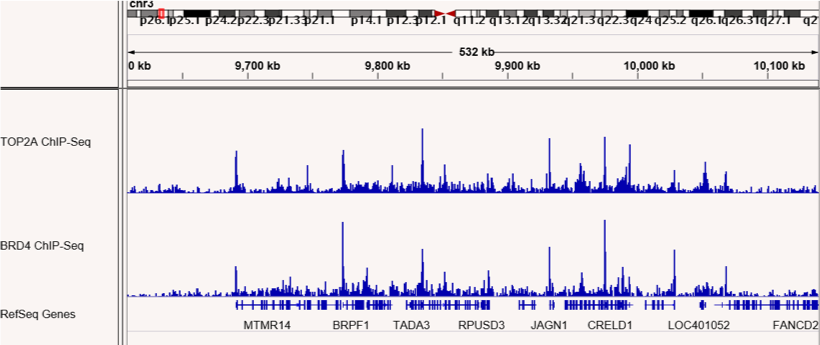
Figures 1 & 2: ChIP-Seq shows co-localization of TOP2A and BRD4 binding sites.
ChIP-Seq was performed to map binding sites for the RIME-identified BRD4 interacting protein, TOP2A. TOP2A and BRD4 binding sites are shown and confirm co-localization at many sites across the genome. The top panel shows sites on chromosome 1 and the bottom panel shows sites on chromosome 3.
References
- †Puc, J. et al. Ligand-Dependent Enhancer Activation Regulated by Topoisomerase-I Activity, Cell. 2015 Oct 8;163(2):520-2.