<< Back to MOTIFvations Blog Home Page
Total Epigenome of the Heart

February 12, 2019
In the spirit of celebrating Valentine’s Day in the nerdiest way possible, we are highlighting an exciting publication that investigated epigenetics in the heart. To characterize the epigenetic changes that accompany aging and heart failure, Ralf Gilsbach and colleagues examined isolated cardiomyocytes from fetal, infantile, adult non-failing, and adult failing hearts.
Getting to the Heart of Epigenetic Analysis – Starting with Sample Prep
Heart tissue is a difficult sample to work with for many types of assays. It is especially difficult to generate high-quality chromatin from the muscle tissue to generate nice ChIP-Seq data. Furthermore, for many studies it will be necessary to investigate only a sub-set of heart tissue cells, such as cardiomyocytes. Isolating cardiomyocytes is not trivial, and the resulting small sample can be difficult to analyze.
However, this is necessary to tease apart cell-specific signatures. In fact, when comparing to a whole heart study, Gilbach and colleagues found that 17% of enhancers detected in whole hearts do not overlap with enhancers detected in cardiomyocytes. Unsurprisingly, these enhancers were located near genes with non-cardiomyocyte functions and were probably detected from other cell types in the heart.
Cell Types in Heart Tissue Change with Age
The issue of other cell types drowning out signal from cardiomyocytes could be exacerbated in adult hearts. After birth, cardiomyocytes lose cell cycle activity while endothelial and mesenchymal cells increasingly proliferate after birth. This is evident when comparing the percentage of the left ventricle composed of cardiomyocytes in fetal and infantile hearts compared to adult (non-failing and failing).
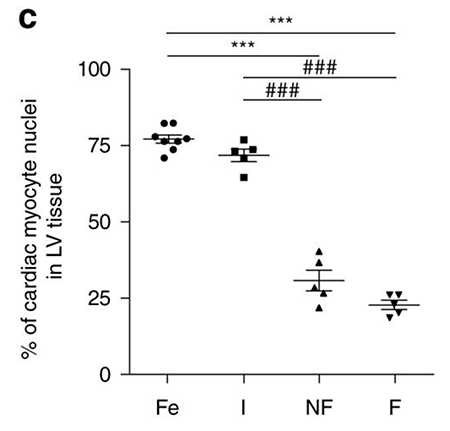
Fetal and infantile left ventricle tissues consist of approximately 75% cardiomyocyctes while adult hearts consist of only 25% cardiomyocytes. Image credit: Gilsbach et al. Nature Communications, Figure 1C.
Evaluating Gene Expression and Epigenetic Profiles in Cardiomyocytes
The researchers next investigated troponin genes in fetal, infantile, adult non-failing, and adult failing cardiomyocytes. Troponin I1 is expressed in fetal cardiomyocyte samples but is epigenetically silenced later in development. Troponin T2 is highly expressed throughout development, and the epigenetic profiles are consistent with the observed abundant expression.
Viewing differential post-translational modifications of histones, methylation of DNA, and expression of RNA provides clues as to the mechanisms of aging and disease in the heart.
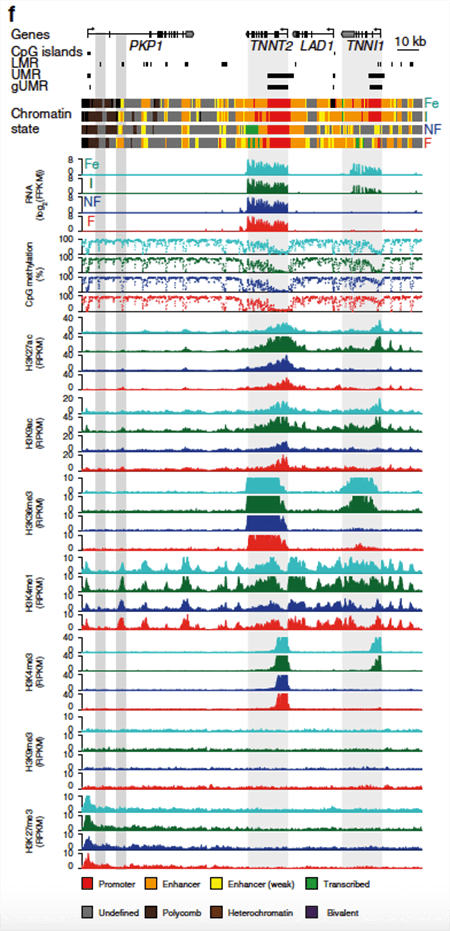
NGS data of gene expression (RNA-Seq), DNA methylation (whole-genome bisulfite sequencing), and histone mark analysis (ChIP-Seq using the Active Motif ChIP-IT High Sensitivity® kit for H3K27ac, H3K9ac, H3K36me3, H3K4me1, H3K4me3, H3K27me3, and H3K9me3) of the troponin I type 1 (TNNI1) and troponin T type 2 (TNNT2) gene region. Image credit: Gilsbach et al. Nature Communications, Figure 1C.
As expected, low levels of DNA CpG methylation correlated with increased levels of active histone marks and increased expression in many genes. Additionally, researchers found that low amounts of DNA methylation were a hallmark of cis-regulatory regions for genes important in cardiomyocyte development, acting either as enhancers or silencers.
Regulatory regions and genes that change expression during development seem to be pre-marked with a histone modification or 5-hmC, which leads to changes in DNA methylation and expression with age.
Interestingly, although DNA methylation shows dynamic changes during development, it remains essentially unchanged in cardiomyocytes in the failing heart relative to the non-failing adult heart. Instead, the researchers found active histone marks correlate with the pathological gene expression associated with heart failure. This distinct form of regulation could be an important clue in the understanding the mechanism and developing treatments for heart failure.
Take Home Message: Cell Type Matters in Epigenomic Analysis
While it’s easier to look at a heterogeneous population of cells in a tissue than isolating the relevant cell types for your experiment, that approach will often not generate the best and most meaningful results.
The development of newer and more sensitive assays, such as the ChIP-IT High Sensitivity® kit, is now making it possible to generate high-quality ChIP-Seq data from as few as 1,000 cells. Improved methods like this are removing previous limitations in sample prep and are allowing researchers to investigate the most appropriate samples in their experiments.
Reference: Gilsbach, R. et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat. Commun. 9: 391 (2018)
Link
<< Back to MOTIFvations Blog Home Page





