ChIC/CUT&RUN Service Overview
Save 20% on CUT&RUN Services. Learn More
CUT&RUN (Cleavage Under Targets & Release Using Nuclease) is an epigenetic method used to investigate the genome-wide distribution of various chromatin-associated proteins and their modifications1-3. CUT&RUN is a derivative of chromatin immunocleavage (ChIC). CUT&RUN is similar to chromatin immunoprecipitation (ChIP), in that it utilizes an antibody to target chromatin-associated marks and proteins, but requires less sample material and less sequencing depths than ChIP.
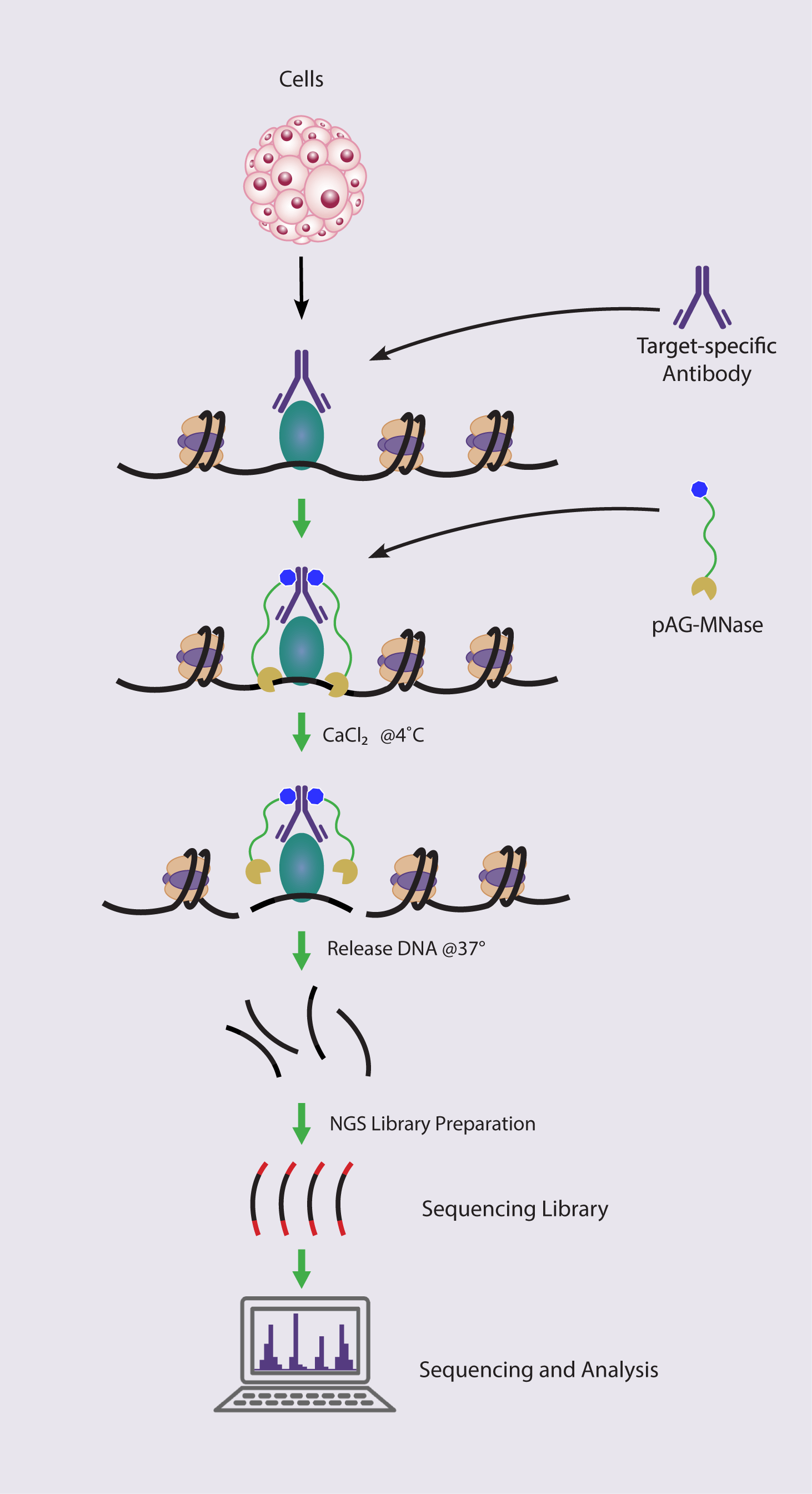
In CUT&RUN, a protein of interest is tagged with an antibody and bound to the chromatin in intact cells. Then, a micrococcal nuclease (MNase) is used to cleave the DNA specifically at the binding sites of the protein of interest. The released fragments are purified, sequenced, and mapped to the reference genome to determine the protein’s binding sites. Unlike ChIP, CUT&RUN does not require crosslinking of the protein to the DNA, which can introduce artifacts.
CUT&RUN is a valuable tool for studying chromatin-associated proteins because it is sensitive, specific, and requires fewer cells than ChIP, making it ideal for identifying binding patterns of chromatin-associated proteins such as transcription factors or histone modifications genome-wide. Chromatin-associated proteins play critical roles in regulating various cellular processes such as gene expression, DNA replication, DNA repair, and cell differentiation. Understanding the binding patterns of these proteins can provide insight into how these cellular processes are regulated.
References:
1. Schmid, M. et al. Mol Cell, 16(1): 147-157 (2004)
2. Skene, P.J. et al. (2017) Elife 6, e21856
3. Skene, P.J. et al. (2018) Nat. Protoc., 13, 1006-1019
Work with the CUT&RUN Experts
Let our team of experts produce high-quality CUT&RUN results from less starting material, saving you time and resources to allow you to focus on the bigger picture.
Using our ChIC/CUT&RUN service is simple. You just submit your samples to Active Motif and receive the analyzed data and publication-ready figures back within a matter of weeks.
To learn more, please submit an Epigenetic Services Information Request.
How the ChIC/CUT&RUN Assay Works
What are the Advantages of Using Active Motif’s ChIC/CUT&RUN Service?
CUT&RUN can be used to map DNA binding proteins including transcription factors, epigenetic readers/writers, RNA polymerase II, or histone modifications, genome-wide, using a fraction of the staring material that is recommended for ChIP-Seq. Having the ability to use less starting material allows for the profiling of histone modifications and transcription factor binding studies in sample types that, due to sample quantities, were not suitable for ChIP-Seq.
CUT&RUN also has the advantage of low background signal such that a lower sequencing depth can be used resulting in high-quality data at a fraction of the cost of ChIP-Seq. To compare with a similar method, CUT&Tag, CUT&RUN is generally more successful in mapping DNA binding proteins than is CUT&Tag. Our ChIC/CUT&RUN Service therefore fills the unmet need of a relatively low-input (compared to ChIP) method to measure genome-wide transcription factor binding.
With our end-to-end ChIC/CUT&RUN Service, you don’t need to worry about protocol optimization or testing multiple antibodies to try to find one that works because we take care of all of that for you. There’s also no need to learn bioinformatics because analysis of the next-generation sequencing data and explanation of the results are included standard.
Active Motif’s end-to-end ChIC/CUT&RUN Service includes:
- Cell preparation
- Concanavalin-A incubation and antibody binding
- Cleavage of antibody-bound chromatin using protein-A/G-MNase
- NGS library preparation and sequencing
- Comprehensive bioinformatics and data analysis
- Delivery of publication-ready figures
ChIC/CUT&RUN Service Data
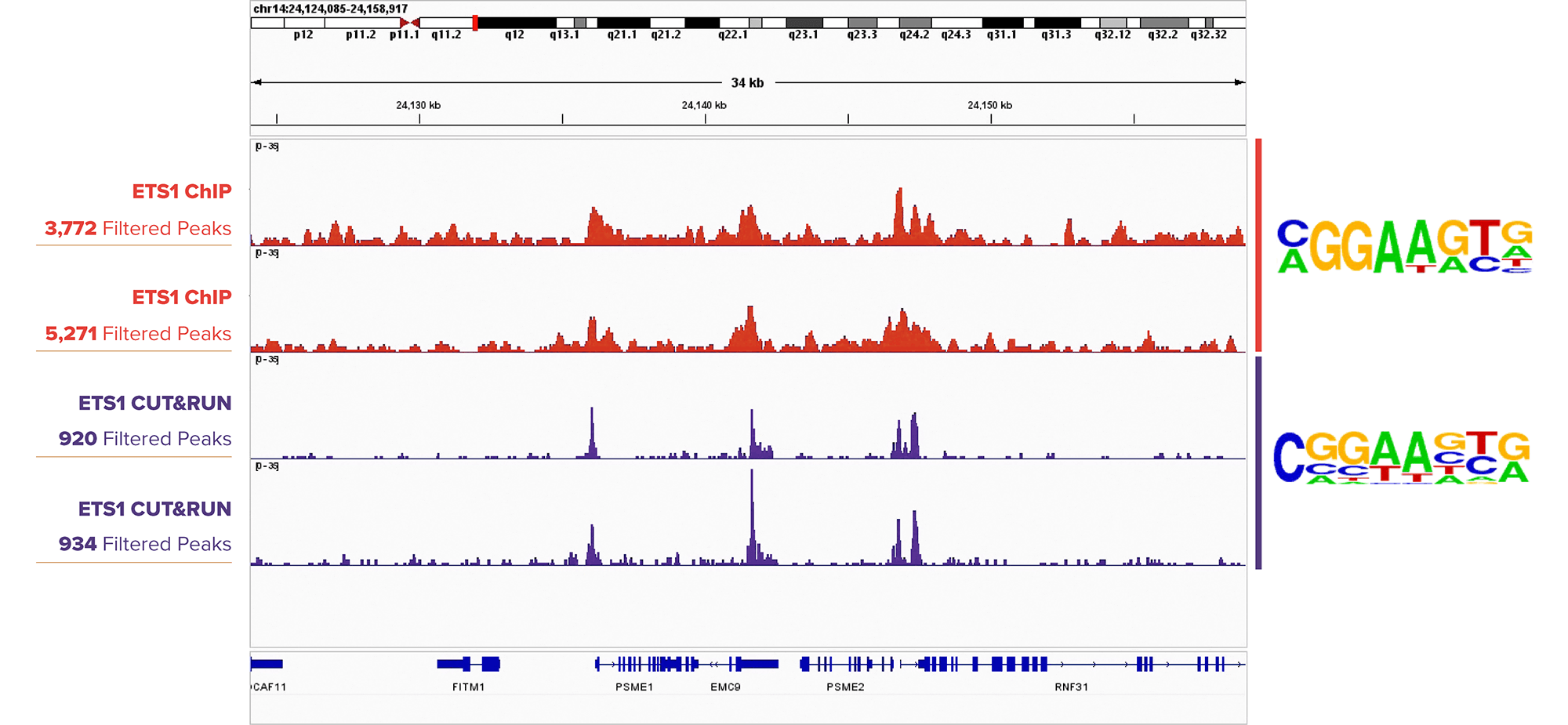
Figure 1. ETS1 CUT&RUN Peak Profiles Are Similar to ETS1 ChIP-Seq Profiles.
IGB browser tracks for ChIP-Seq (top two tracks) in DIPTG cells and CUT&RUN (bottom two tracks) in K562 cells assays targeting ETS1 (Cat. No. 39580). Sequence logos from Homer analysis (to the right of the tracks) show motif enrichment for the ETS1 motif in both sample sets.
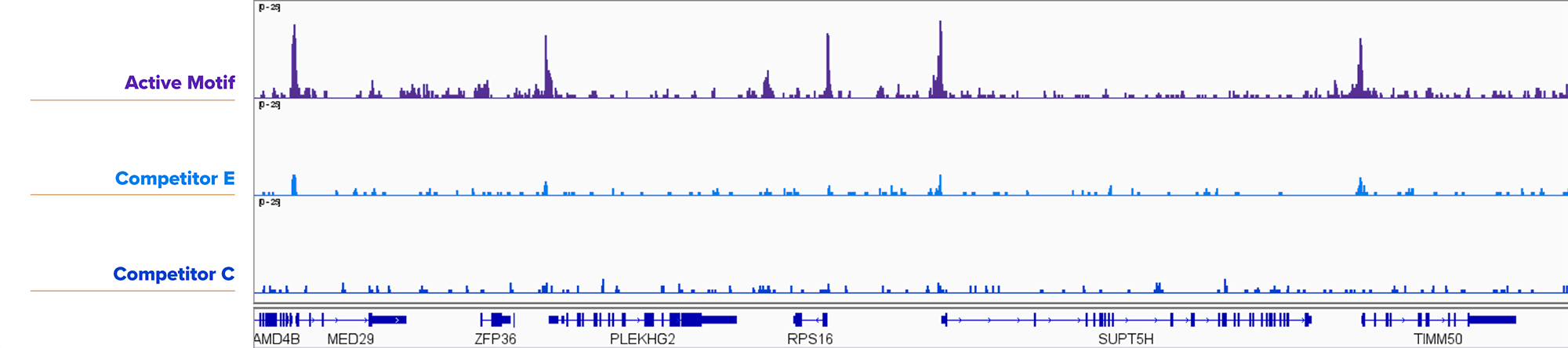
Figure 2. Active Motif’s ChIC/CUT&RUN Assay Kit is Compatible with 25,000 Nuclei for Transcription Factors.
25,000 fresh K562 nuclei were assayed using Active Motif’s CUT&RUN Assay Kit or Competitor E or Competitor C CUT&RUN Assay Kits using Active Motif's YY1 antibody (Cat. No. 61980). Active Motif’s CUT&RUN Assay Kit yielded more robust results than the competition.
ChIC/CUT&RUN Service Documents
ChIC/CUT&RUN Service Sample Submission Portal
Our online sample submission portal allows you to easily upload your service project samples and track your project status. Follow the sample submission instructions in the portal to ensure that all your samples arrive at Active Motif in the best possible condition and properly associated with your project.
You might also be interested in:
| Name | Cat No. | Price | |
|---|---|---|---|
| ChIC/CUT&RUN Service | 25160 | Get Quote | |