Bisulfite Conversion Overview
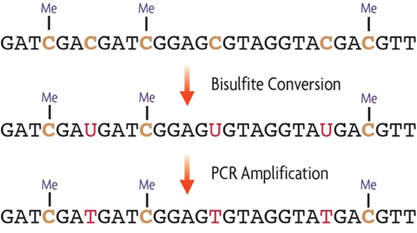
Bisulfite Conversion is a method used to make it easy to distinguish methylated from unmethylated cytosines in genomic DNA at single base resolution.
DNA is first denatured (made single-stranded) and then treated with sodium bisulfite. Sodium bisulfite selectively changes unmethylated cytosines into uracils through deamination, while leaving methylated cytosines (both 5-methylcytosine and 5-hydroxymethylcytosine) unchanged. By then amplifying the treated DNA with PCR, uracils are further converted to thymines. At this point all nucleotides that were originally unmethylated cytosines become thymines while those that were methylated cytosines remain cytosines. The result is a clear nucleotide difference between methylated and unmethylated cytosines that can be easily identified by comparison to the original reference genome.
Bisulfite Conversion Services
Active Motif’s Reduced Representation Bisulfite Sequencing (RRBS) Service provides single base methylation data for up to 5 million CpGs with a sequencing depth greater than 30 million reads per sample. The service is an end-to-end assay and includes DNA purification, digestion, bisulfite conversion, library preparation, DNA sequencing, and bioinformatics analysis. To learn more about how RRBS works see below.
Active Motif’s Targeted Next-Gen Bisulfite Sequencing Service is a single base-pair, high-throughput solution for targeting a handful of regions of interest. The service is an end-to-end assay and includes bisulfite conversion, primer design and testing, PCR amplification, library preparation, DNA sequencing and bioinformatics analysis. To learn more about how targeting bisulfite sequencing works see below.
Applications for Bisulfite Conversion
Reduced Representation Bisulfite Sequencing (RRBS)
Reduced Representation Bisulfite Sequencing (RRBS) uses restriction enzymes together with DNA size selection to greatly reduce the percentage of the genome that must be sequenced. DNA is first digested using methylation-insensitive restriction enzymes (commonly MspI) which generate fragments containing CpG dinucleotides at the ends. Fragments are then adapter ligated and size selected so that CpG rich regions of DNA are captured. Enriched fragments can then be bisulfite converted and amplified to reveal methylated cytosines and sequenced. RRBS captures 80-85% of CpG islands and 50-60% of human promoters while sequencing only 3% of the genome.
Active Motif’s Reduced Representation Bisulfite Sequencing Service provides single base methylation data for up to 5 million CpGs with a sequencing depth greater than 30 million reads per sample. The service is an end-to-end assay and includes DNA purification, digestion, bisulfite conversion, library preparation, DNA sequencing, and bioinformatics analysis.
Advantages: Can sequence regions dense in CpG methylation genome wide at single-base resolution for lower cost than WGBS.
Disadvantages: Restriction enzymes are biased. There is no coverage of non CpG rich areas. Misses 85-90% of CpGs in the genome.
Best Uses: Good method to scan for changes in CpG rich regions like promotors which are commonly abnormally methylated in diseases such as cancer.
Targeted Bisulfite Sequencing
Targeted bisulfite sequencing allows for very accurate analysis of methylation at single nucleotide resolution of specific regions of interest. It begins with bisulfite conversion of DNA and then uses primers to PCR amplify targeted loci. These amplicons then undergo library construction and are sequenced. By limiting sequencing to a small number of multiplexed amplicons, high sequencing depth of the cytosines is achieved for those regions at low cost.
Active Motif’s Targeted Next-Gen Bisulfite Sequencing Service is a single base-pair, high-throughput solution for targeting a handful of regions of interest. The service is an end-to-end assay and includes bisulfite conversion, primer design and testing, PCR amplification, library preparation, DNA sequencing and bioinformatics analysis.
Advantages: High sequencing depth at low cost for regions of interest.
Disadvantages: Primers must be designed for every region.
Best Uses: Like MSP it’s a good method when interested in a small number of regions of interest.
Methylation-specific PCR (MSP)
Methylation-specific PCR (MSP) is used to probe targeted regions for methylation after bisulfite conversion. It uses two different methylation specific primer sets, one which complements methylated DNA and one which complements unmethylated DNA. PCR can be performed to amplify the targeted DNA and PCR products run on a gel to visualize which primer set produced more amplified product. Alternatively, a TaqMan® probe approach can be used where methylation specific fluorescent probes are bound to DNA and non-methylation specific PCR primers are used to amplify the region. Upon amplification, bound probes are degraded by Taq polymerase, so that light emitted from the probe increases. An increase in the signal for either the methylated or unmethylated specific probe indicates the methylation state of the DNA.
Advantages: Low cost, fast execution.
Disadvantages: Can be difficult to design primers as they require at least five thymines.
Best Uses: Best suited for assessing methylation in a small number of regions of interest.
Whole Genome Bisulfite Sequencing (WGBS)
Often referred to as the “gold standard” for analysis of DNA methylation, Whole Genome Bisulfite Sequencing (WGBS) provides the greatest coverage and resolution of any DNA methylation analysis technique. Genomic DNA is first isolated and undergoes bisulfite conversion. The DNA is then adapter ligated and used to prepare a sequencing library. Following sequencing, DNA is bioinformatically analyzed to reveal methylated cytosines across the entire genome. The drawback to such comprehensive coverage is that the DNA is costly to sequence and complex to analyze.
Advantages: Single-nucleotide resolution of DNA methylation across the entire genome.
Disadvantages: Expensive and time-consuming sequencing analysis. Not practical for high throughput.
Best Uses: Ideal when interested in low-CpG-density regions, intergenic “gene deserts”, or partially methylated domains and distal regulatory elements, that would be missed by RRBS.
Methylation Array
Methylation arrays allow for quantification of methylation at selected cytosines in multiplexed samples on a chip. DNA is first bisulfite treated and PCR amplified to reveal the methylation pattern of the DNA. It is then combined with DNA probes which target specific methylation sites. One method used two probes for each methylation site, one complimentary to the methylated state and the other to the unmethylated state of the cytosine. Another method binds a single probe to each site and distinguishes between methylation states based on a single base extension over the methylation site. Differently tagged nucleotides reveal levels of methylation from a fluorescent signal that is specific to the unmethylated or methylated state at each site.
Advantages: Easy to use. Reproducible. Can be used with FFPE samples.
Disadvantages: Arrays work for human or mouse DNA only. Many methylation sites are included in the assay. Covers most CpG islands (>96%) but less than 2% of CpG sites in human genome.
Best Uses: Great for clinical screening of samples for aberrant methylation differences in CpG islands.

