The ChIP-IT® qPCR Analysis Kit was designed to simplify ChIP qPCR analysis and data interpretation. The kit includes standard curve DNA, a standard curve primer pair, positive and negative control qPCR primer sets for human and mouse samples and an analysis spreadsheet that automatically normalizes and graphs the data. The normalization calculations take into consideration the amount of chromatin that was used in the ChIP reaction, the resuspension volume of the ChIP DNA and the primer efficiencies to introduce a level of consistency in the data output that enables a reliable comparison of many samples across multiple experiments. Based on the consistency of the data output, Active Motif is able to make specific recommendations in regards to data interpretation so that customers can reliably assess the quality of their ChIP DNA. We recommend using Active Motif's ChIP-IT High Sensitivity® Kit to perform the chromatin immunoprecipitation reactions.
Please click on the link to download a copy of the ChIP-IT® qPCR Analysis Spreadsheet.
| Name | Format | Cat No. | Price | |
|---|---|---|---|---|
| ChIP-IT® qPCR Analysis Kit | 10 rxns | 53029 | $125 | Buy |
ChIP-IT® qPCR Analysis Kit Advantages
- Simplifies qPCR data analysis to enable a faster, more reliable interpretation of data
- All calculations are included in a simple Excel sheet in which data from your qPCR machine can be entered directly resulting in automatic data normalization and graph generation
- Normalized data can be compared across multiple sample types and experiments
- Includes human and mouse positive control and negative control qPCR primer sets
- Recommendations for acceptable data values allows users to evaluate the quality of the ChIP reaction before proceeding with downstream applications such as ChIP-Seq
How does it work?
Using the ChIP DNA obtained from the ChIP-IT High Sensitivity Kit, qPCR reactions are set up for the provided DNA Standards, the ChIP DNA samples and Input DNA. An example 96-well plate qPCR set up using six different ChIP DNA samples and four different PCR primer pairs is shown below. Each ChIP and Input reaction is tested in triplicate using the provided species-specific negative control primer set and a positive control primer. Since the DNA mass in each standard is known, the number of copies of the genome present in each standard can be calculated (assuming that each cell contains 6.6 pg of DNA). The primer efficiency is calculated for each primer set and the qPCR values for the ChIP reactions are normalized against primer efficiency, the amount of chromatin used in the ChIP reaction and the resuspension volume of the ChIP DNA. To see a complete explanation of the data calculations and the recommendations for data interpretation, please download a copy of the product manual under the Documents tab.
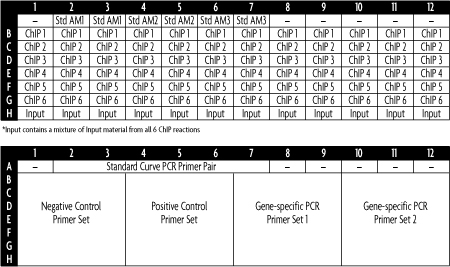
Figure 1: qPCR plate set up for use with the ChIP-IT qPCR Analysis Kit.
This example 96-well plate set up shows how the ChIP-IT qPCR Analysis Kit can be used to compare six different ChIP DNA samples with 4 different qPCR primer sets. Only one standard curve is needed for the entire plate.
Data Interpretation
Active Motif has done an extensive evaluation of ChIP DNA quality using the ChIP-IT qPCR Analysis Kit and normalization strategy. Based on these results we are able to provide specific recommendations for assessing ChIP DNA quality. These recommendations are based on the use of the ChIP-IT High Sensitivity Kit (Catalog No. 53040) to perform the chromatin immunoprecipitation reactions. If another method was used to perform the ChIP reactions, the interpretation results listed below may not apply. Below are a few example recommendations. A complete description can be found in the product manual under the Documents tab.
- Negative control primers should give values of less than 2 Binding events detected per 1,000 cells. Values higher than 2 could indicate a poor quality ChIP reaction
- ChIP reactions using antibodies against high abundance targets, such as histone modifications, will routinely give values between 100 and 1,000 Binding events detected per 1,000 cells.
- ChIP reactions using antibodies against lower abundance targets, such as transcription factors, will routinely give values between 10 and 100 Binding events detected per 1,000 cells.
- ChIP reactions that give values less than 5 Binding events detected per 1,000 cells are usually not of high quality. The ChIP reaction should be repeated or a different antibody should be used.
- A successful ChIP reaction will have a minimum of 5-fold higher values with the positive control primers as compared to the negative control primers.
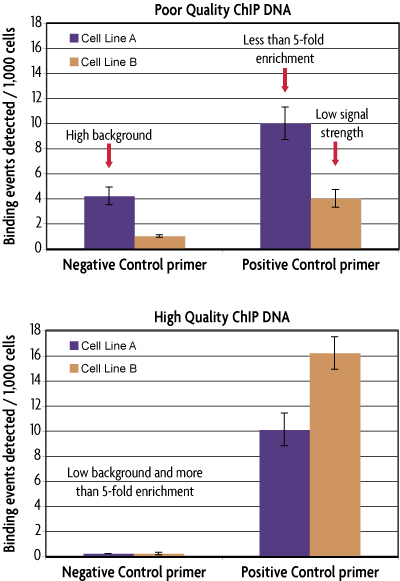
Figure 1: The ChIP-IT qPCR Analysis Kit and normalization strategy provide an effective means of evaluating ChIP DNA quality.
The ChIP-IT qPCR Analysis Kit manual provides recommendations for evaluating the quality of ChIP reactions. The top image shows two examples of ChIP reactions that do not meet the recommended criteria for quality ChIP DNA. ChIP DNA from Cell Line A shows high background and less than 5-fold enrichment, while ChIP DNA from Cell Line B shows low signal strength. The bottom image show ChIP DNA that is considered high quality as both samples show low background and greater than 5-fold enrichment.
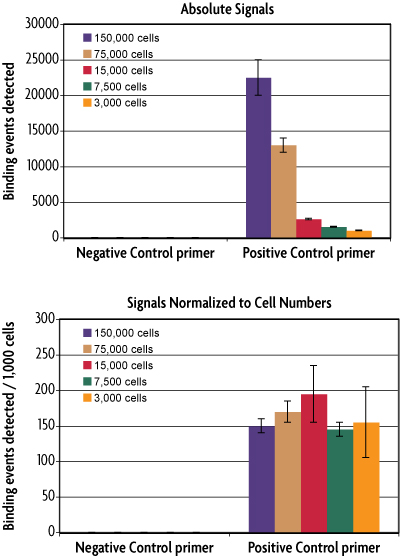
Figure 2: The ChIP-IT qPCR Analysis Kit enables direct comparison of ChIP efficiency.
Figure 2: The ChIP-IT qPCR Analysis Kit enables direct comparison of ChIP efficiency.
ChIP was performed using an RNA pol II antibody with the ChIP-IT High Sensitivity Kit. Different amounts of chromatin were used in each ChIP reaction ranging from 3,000 cell equivalents to 150,000 cell equivalents. In the top image normalization was performed independent of starting chromatin amount, taking only primer efficiency and resuspension volume into account. In the bottom image, using Active Motif's ChIP-IT qPCR Analysis Kit, starting amount of chromatin is accounted for. This data demonstrates the dynamic range of the ChIP-IT High Sensitivity Kit and the accuracy of the normalization strategy in the ChIP-IT qPCR Analysis Kit.
Contents & Storage
The ChIP-IT® qPCR Analysis Kit is shipped on dry ice and contains reagents with multiple storage temperatures inside. Please store each component at the temperature indicated below.
- DNA Standard AM1; Store at -20°C
- DNA Standard AM2; Store at -20°C
- DNA Standard AM3; Store at -20°C
- Standard Curve Primer Pair; Store at -20°C
- Human Negative Control Primer Set 1; Store at -20°C
- Human Positive Control Primer Set GAPDH-2; Store at -20°C
- Mouse Negative Control Primer Set 1; Store at -20°C
- Mouse Positive Control Primer Set Gapdh-2; Store at -20°C
- 2 mM Tris-HCl, pH 8.0; Store at RT
Search our database of customer publications that have used our ChIP-IT® qPCR Analysis Kit.


