Single-Cell and Single-Nucleus RNA-Seq Overview
Single-Cell RNA-Seq (scRNA-Seq) enables transcriptome analysis at the single-cell level. Understanding the transcriptome profile at single-cell resolution can help deconvolute gene expression in heterogenous populations.
While most of the messenger RNA in a cell typically resides in the cytosol, the most recently expressed transcripts can be found in the nucleus. When using viable intact cells as a starting material we measure gene expression from the mRNA in the whole cell. But, when using frozen tissue as a starting material, it is much easier to isolate intact nuclei than intact cells. For frozen tissue we offer Single-Nucleus RNA-Seq (snRNA-Seq).
While Single-Nucleus RNA-Seq results are admittedly different from Single-Cell RNA-Seq results, the conclusions of the above publications show that there is an almost complete overlap of global gene expression detected between the two methods. The consensus is that Single-Nucleus RNA-seq sufficiently identifies cell types within a heterogeneous population based on transcriptomal differences.
Several publications have compared the data obtained from Single-Cell RNA-Seq to Single-Nucleus RNA-Seq:
What are the advantages of using scRNA-Seq or snRNA-Seq?
scRNA-Seq or snRNA-Seq can be used to identify cell subpopulations with different transcriptome profiles within complex samples, eliminating the need for isolation strategies like FACS or magnetic sorting that could alter the biology of the sample due to sample manipulation. For example:
- Identifying novel cell subpopulations that are responsible for response to drug treatments (responders vs. resistant cells)
- Identifying subpopulations of cells with variations in gene expression that can provide insight into developmental trajectories (for example, brain development, T-helper cell development, B-cell differentiation)
Active Motif’s end-to-end scRNA/snRNA-Seq Service includes:
- Cell or nuclei preparation
- Sample processed using 10X Genomics Chromium platform
- Library generation
- Sequencing
- Bioinformatic analysis
Single-Cell and Single-Nucleus RNA-Seq Data
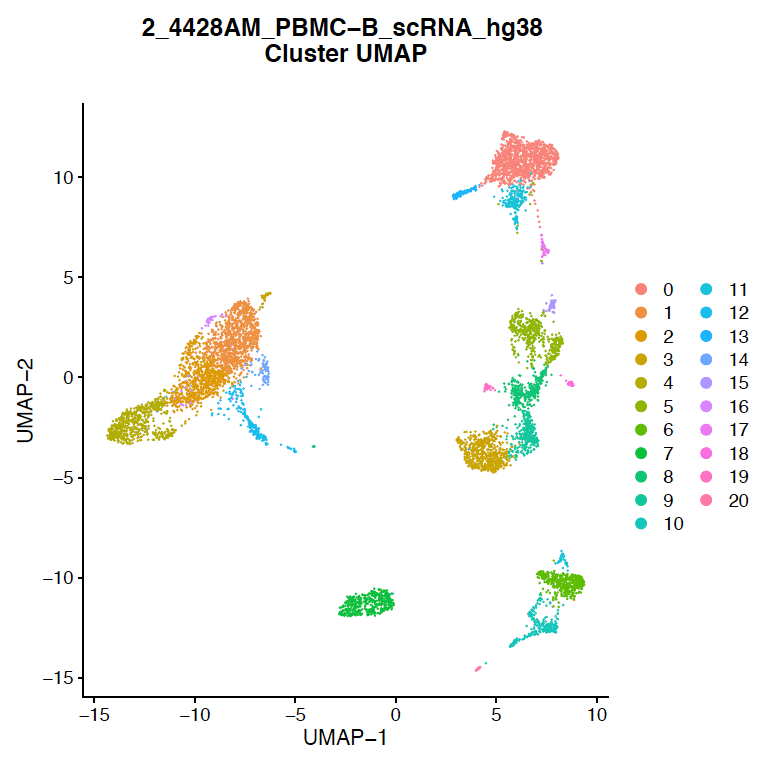
Figure 1: Identify unique subpopulations of cells within a single sample.
Single-Cell RNA-Seq data generated from human PBMCs. Each color-coded cluster on the UMAP plot represents populations of cells that have the same gene expression profile. 20 refined clusters were identified.
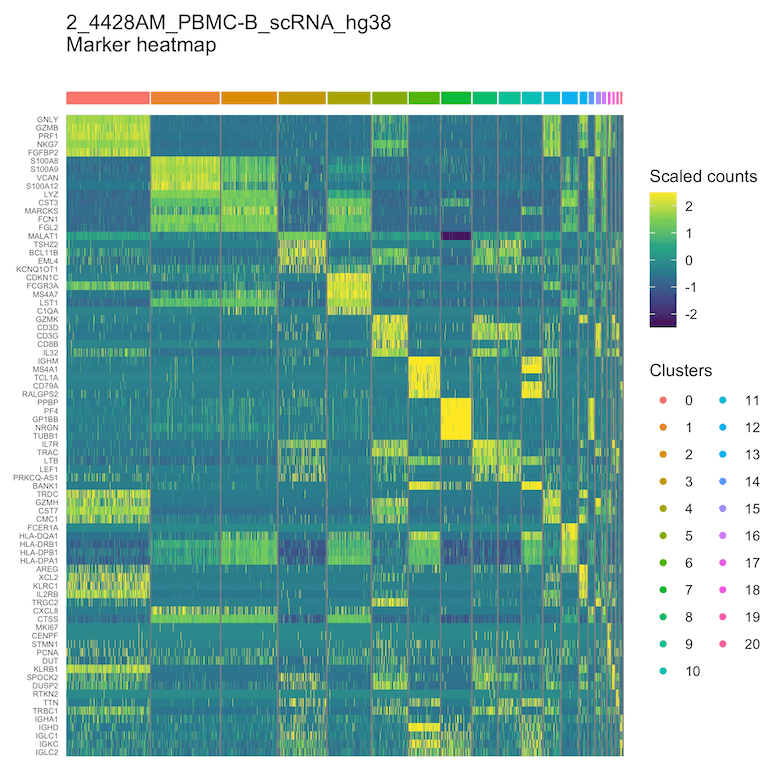
Figure 2: Heatmap of differentially expressed genes per cluster.
Genes that are differentially expressed in each cluster are displayed as a heatmap.
Single-Cell and Single-Nucleus RNA-Seq FAQ
What is single-cell RNA-Seq?
How is scRNA-Seq different from bulk RNA-Seq?
Is this service only offered for human samples?
Is this service available for tissue samples?
Is scRNA-Seq compatible with FFPE samples?
How many cells are required for Active Motif’s scRNA-Seq Service?
What is the read depth for Active Motif’s scRNA-Seq Service?
Single-Cell and Single-Nucleus RNA-Seq Documents
Single-Cell and Single-Nucleus RNA-Seq Sample Submission Portal
Our online sample submission portal allows you to easily upload your service project samples and track your project status. Follow the sample submission instructions in the portal to ensure that all your samples arrive at Active Motif in the best possible condition and properly associated with your project.